BioPhys - Computational Biology
BioPhys - Computational Biology
Componenti
- Caselle Michele (Coordinatore/Coordinatrice)
- Osella Matteo (Professore/Professoressa Associato/a)
- Filippo Valle (Dottorando/a)
- Marta Biondo (Dottorando/a)
Contatti
Settore ERC
Attività
The main goal of the BioPhys network is the study of problems and systems of Biological interest with the tools and ideas typical of theoretical physics. Our main focus is in Molecular Biology, a research context which has seen an impressive development in these last years and is nowadays of central relevance, both for the deep scientific questions which it raises and for the large number of potential biomedical applications.
In this framework, our research project is organized in three main directions:
1. Numerical simulations of systems of biological interest. (protein folding, biomolecules aggregation, protein-protein interactions...)
2. Neural Networks and related systems. (Study of Brain connectivity, emergence of spatio-temporal patterns....)
3. Regulatory Networks (Gene Regulatory Networks, Immune Networks, Chromatin organization....)
The common theme and the unifying setting of these research lines is the use of theoretical physics methods to model and analyse these problems. In particular methods borrowed from the statistical mechanics of disordered systems (Spin Glasses, Hopfield model...); computational tools typical of the simulation of many-body physical systems (Montecarlo Simulations, Molecular Dynamics,...); advanced mathematical tools (Graph theory, Network Theory, Stochastic Equations....)
The most relevant aspect of our research network is the strong interaction of all the units with biologist. All the problems that we address stem from precise biological questions to which we try to give quantitative and testable answers.
2012-2015 "Grant di Ateneo" GeneRNet
"Computational and experimental approaches to identify and model biologically relevant Gene Regulatory Networks"
2022-2025 CRT I TORNATA 2022 - "GENPHYS: STATISTICAL PHYSICS FOR GENOMIC DATA MINING"
Prodotti della ricerca
- Protein degradation sets the fraction of active ribosomes at vanishing growth
2022-01-01 Calabrese L.; Grilli J.; Osella M.; Kempes C.P.; Lagomarsino M.C.; Ciandrini L. https://iris.unito.it/handle/2318/1885184 - A 3D transcriptomics atlas of the mouse nose sheds light on the anatomical logic of smell
2022-01-01 Ruiz Tejada Segura, Mayra L.; Abou Moussa, Eman; Garabello, Elisa; Nakahara, Thiago S.; Makhlouf, Melanie; Mathew, Lisa S.; Wang, Li; Valle, Filippo; Huang, Susie S.Y.; Mainland, Joel D.; Caselle, Michele; Osella, Matteo; Lorenz, Stephan; Reisert, Johannes; Logan, Darren W.; Malnic, Bettina; Scialdone, Antonio; Saraiva, Luis R. https://iris.unito.it/handle/2318/1850917 - The Dynamics of Aerotaxis in a Simple Eukaryotic Model
2021-01-01 Biondo M.; Panuzzo C.; Ali S.M.; Bozzaro S.; Osella M.; Bracco E.; Pergolizzi B. https://iris.unito.it/handle/2318/1841854 - Initial cell density encodes proliferative potential in cancer cell populations
2021-01-01 Enrico Bena C.; Del Giudice M.; Grob A.; Gueudre T.; Miotto M.; Gialama D.; Osella M.; Turco E.; Ceroni F.; De Martino A.; Bosia C. https://iris.unito.it/handle/2318/1803600 - Concurrent processes set E. coli cell division
2018-01-01 Micali, Gabriele; Grilli, Jacopo; Osella, Matteo; Cosentino Lagomarsino, Marco https://iris.unito.it/handle/2318/1685876 - Dissecting the Control Mechanisms for DNA Replication and Cell Division in E. coli
2018-01-01 Micali, Gabriele; Grilli, Jacopo; Marchi, Jacopo; Osella, Matteo; Cosentino Lagomarsino, Marco https://iris.unito.it/handle/2318/1685879 - Relevant parameters in models of cell division control
2017-01-01 Grilli, Jacopo; Osella, Matteo; Kennard, Andrew S; Lagomarsino, Marco Cosentino https://iris.unito.it/handle/2318/1685884 - Step by Step, Cell by Cell: Quantification of the Bacterial Cell Cycle
2017-01-01 Osella, Matteo; Tans, Sander J; Cosentino Lagomarsino, Marco https://iris.unito.it/handle/2318/1685885 - Emergent statistical laws in single-cell transcriptomic data
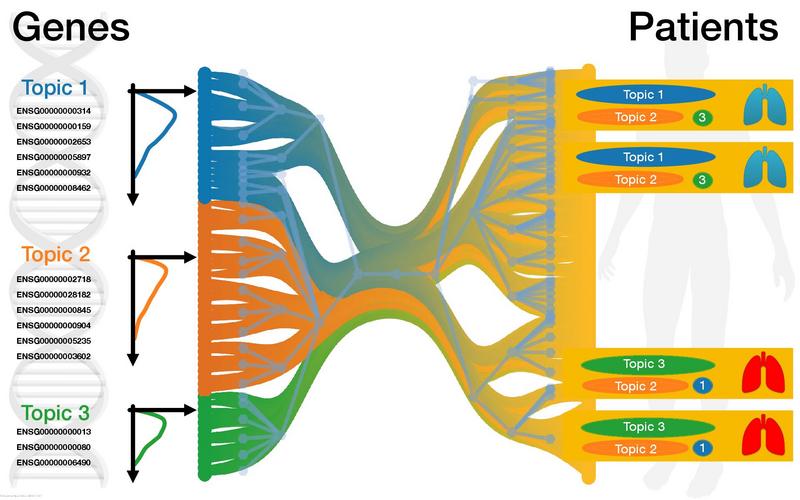
2023-01-01 Lazzardi, Silvia; Valle, Filippo; Mazzolini, Andrea; Scialdone, Antonio; Caselle, Michele; Osella, Matteo https://iris.unito.it/handle/2318/1906810 - Multiomics Topic Modeling for Breast Cancer Classification
2022-01-01 Valle F.; Osella M.; Caselle M. https://iris.unito.it/handle/2318/1850916 - The impact of whole genome duplications on the human gene regulatory networks
2021-01-01 Mottes F.; Villa C.; Osella M.; Caselle M. https://iris.unito.it/handle/2318/1841857 - A Topic Modeling Analysis of TCGA Breast and Lung Cancer Transcriptomic Data
2020-01-01 Valle, Filippo; Osella, Matteo; Caselle, Michele https://iris.unito.it/handle/2318/1767492 - A Model-Driven Quantitative Analysis of Retrotransposon Distributions in the Human Genome
2020-01-01 Riba, Andrea; Fumagalli, Maria Rita; Caselle, Michele; Osella, Matteo https://iris.unito.it/handle/2318/1767481 - Investigating the epi-miRNome: Identification of epi-miRNAs using transfection experiments
2019-01-01 Reale E.; Taverna D.; Cantini L.; Martignetti L.; Osella M.; De Pitta C.; Virga F.; Orso F.; Caselle M. https://iris.unito.it/handle/2318/1724081 - Biophysical Analysis of miRNA-Dependent Gene Regulation
2018-01-01 Riba, Andrea; Osella, Matteo; Caselle, Michele; Zavolan, Mihaela https://iris.unito.it/handle/2318/1685895 - Statistics of Shared Components in Complex Component Systems
2018-01-01 Mazzolini, Andrea; Gherardi, Marco; Caselle, Michele; Cosentino Lagomarsino, Marco; Osella, Matteo https://iris.unito.it/handle/2318/1675883 - Heaps' law, statistics of shared components, and temporal patterns from a sample-space-reducing process
2018-01-01 Mazzolini, Andrea; Colliva, Alberto; Caselle, Michele; Osella, Matteo https://iris.unito.it/handle/2318/1685855 - Modelling the evolution of transcription factor binding preferences in complex eukaryotes
2017-01-01 Rosanova, Antonio; Colliva, Alberto; Osella, Matteo; Caselle, Michele https://iris.unito.it/handle/2318/1650227 - Stochastic timing in gene expression for simple regulatory strategies
2017-01-01 Co, Alma Dal; Lagomarsino, Marco Cosentino; Caselle, Michele; Osella, Matteo https://iris.unito.it/handle/2318/1628379 - Interplay of microRNA and epigenetic regulation in the human regulatory network.
2014-01-01 Osella M;Riba A;Testori A;Corà D;Caselle M https://iris.unito.it/handle/2318/151515 - Gene autoregulation via intronic microRNAs and its functions.
2012-01-01 Bosia C; Osella M; El Baroudi M; Corà D; Caselle M. https://iris.unito.it/handle/2318/130300 - The role of incoherent microRNA-mediated feedforward loops in noise buffering
2011-01-01 Caselle M; Osella M; Bosia C; El Baroudi M; Cora D https://iris.unito.it/handle/2318/89862 - Gene autoregulation via intronic microRNAs and its functions
2011-01-01 El Baroudi M; Bosia C; Osella M; Cora' D; Caselle M https://iris.unito.it/handle/2318/89863 - A Curated Database of miRNA Mediated Feed-Forward Loops Involving MYC as Master Regulator.
2011-01-01 El Baroudi M; Corà D; Bosia C; Osella M; Caselle M. https://iris.unito.it/handle/2318/87274 - The Role of Incoherent MicroRNA-Mediated Feedforward Loops in Noise Buffering
2011-01-01 Matteo Osella; Carla Bosia; Davide Cora'; Michele Caselle https://iris.unito.it/handle/2318/86735 - Entropic contributions to the splicing process
2009-01-01 M. Osella; M. Caselle https://iris.unito.it/handle/2318/73143 - Identification of microRNA clusters cooperatively acting on epithelial to mesenchymal transition in triple negative breast cancer
2019-01-01 Cantini L.; Bertoli G.; Cava C.; Dubois T.; Zinovyev A.; Caselle M.; Castiglioni I.; Barillot E.; Martignetti L. https://iris.unito.it/handle/2318/1712179 - Molecular inverse comorbidity between alzheimer’s disease and lung cancer: New insights from matrix factorization
2019-01-01 Greco A.; Valle J.S.; Pancaldi V.; Baudot A.; Barillot E.; Caselle M.; Valencia A.; Zinovyev A.; Cantini L. https://iris.unito.it/handle/2318/1712181 - Hope4Genes: a Hopfield-like class prediction algorithm for transcriptomic data
2019-01-01 Cantini L.; Caselle M. https://iris.unito.it/handle/2318/1712180 - A review of computational approaches detecting microRNAs involved in cancer
2017-01-01 Cantini, L; Caselle, M; Forget, A; Zinovyev, A; Barillot, E; Martignetti, L https://iris.unito.it/handle/2318/1633746 - MicroRNA–mRNA interactions underlying colorectal cancer molecular subtypes
2015-01-01 Cantini, Laura; Isella, Claudio; Petti, Consalvo; Picco, Gabriele; Chiola, Simone; Ficarra, Elisa; Caselle, Michele; Medico, Enzo https://iris.unito.it/handle/2318/1529830 - Detection of gene communities in multi-networks reveals cancer drivers
2015-01-01 Cantini, Laura; Medico, Enzo; Fortunato, Santo; Caselle, Michele https://iris.unito.it/handle/2318/1547072